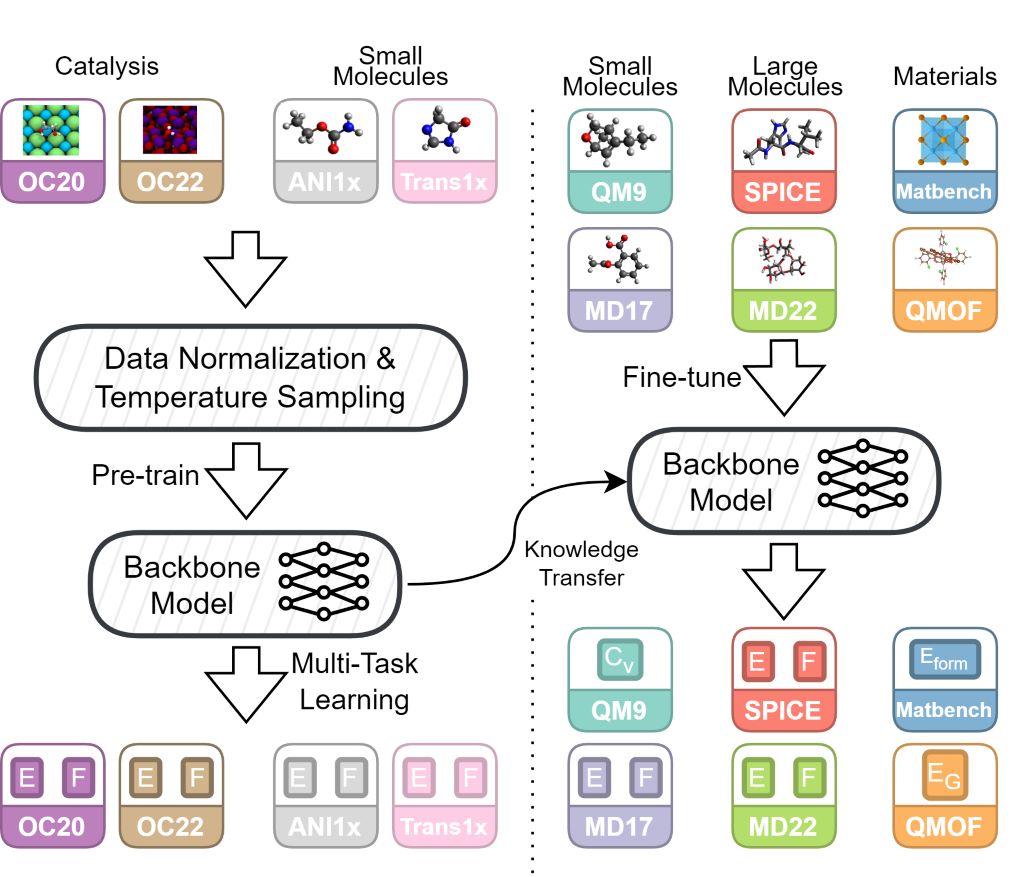
Foundation models have been transformational in machine learning fields such as natural language processing and computer vision. Similar success in atomic property prediction has been limited due to the challenges of training effective models across multiple chemical domains. To address this, we introduce Joint Multi-domain Pre-training (JMP), a supervised pre-training strategy that simultaneously trains on multiple datasets from different chemical domains, treating each dataset as a unique pre-training task within a multi-task framework. Our combined training dataset consists of ~120M systems from OC20, OC22, ANI-1x, and Transition-1x. We evaluate performance and generalization by fine-tuning over a diverse set of downstream tasks and datasets including: QM9, rMD17, MatBench, QMOF, SPICE, and MD22. JMP demonstrates an average improvement of 59% over training from scratch, and matches or sets state-of-the-art on 34 out of 40 tasks. Our work highlights the potential of pre-training strategies that utilize diverse data to advance property prediction across chemical domains, especially for low-data tasks.
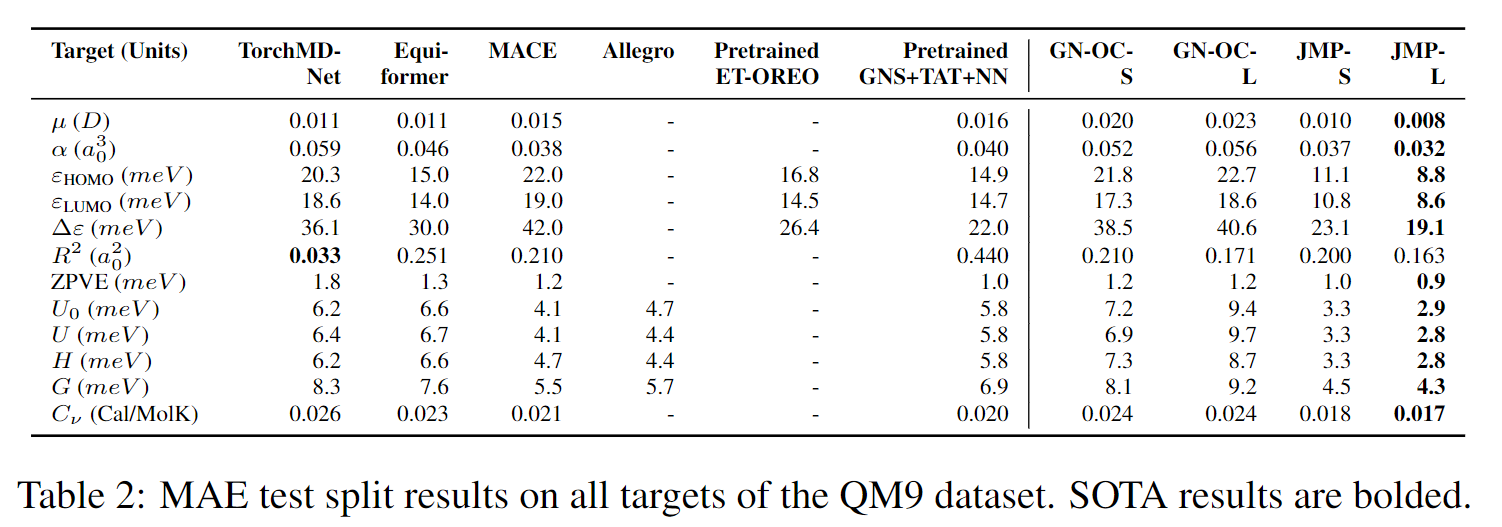
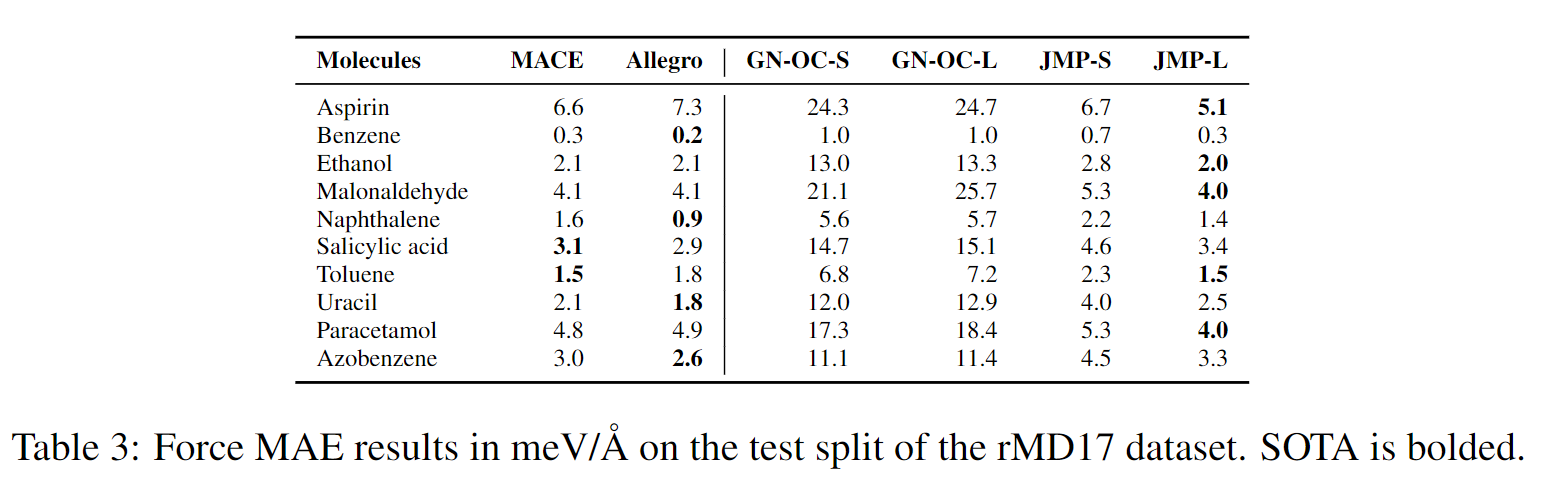
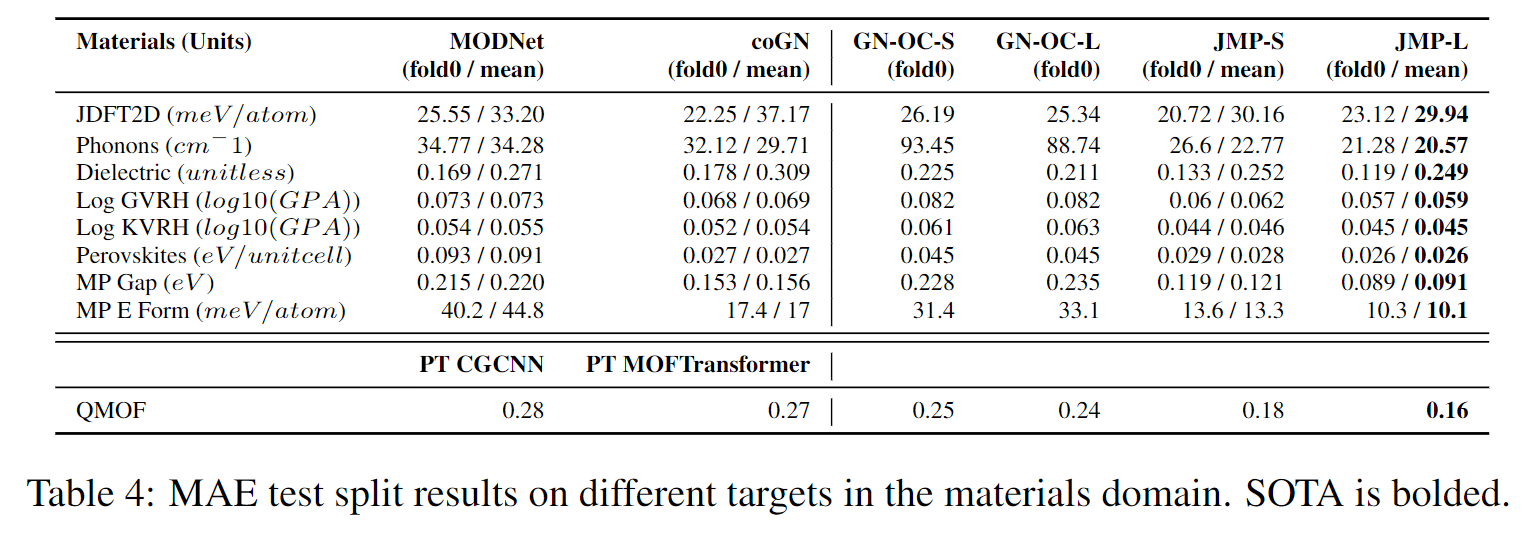
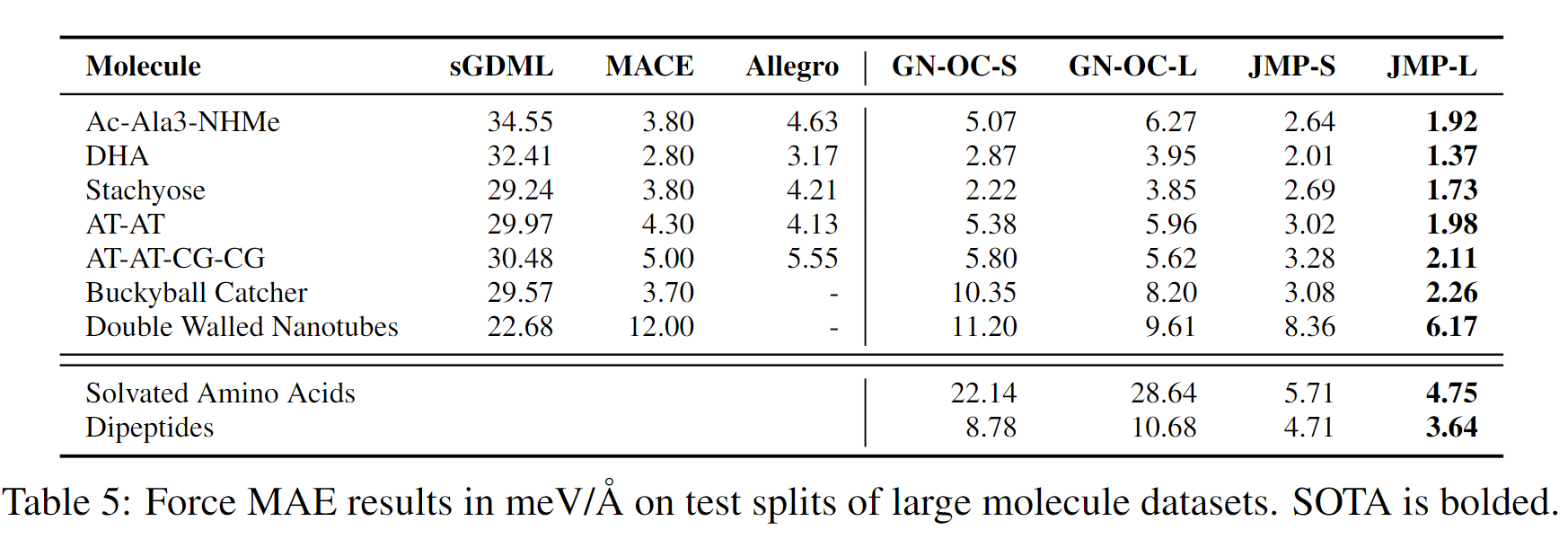
JMP-L consistently outperforms the scratch variant across all tasks, with an average improvement of 59%. JMP-L also matches or sets state-of-the-art on 34 out of 40 tasks, demonstrating the effectiveness of pre-training on diverse data for atomic property prediction.
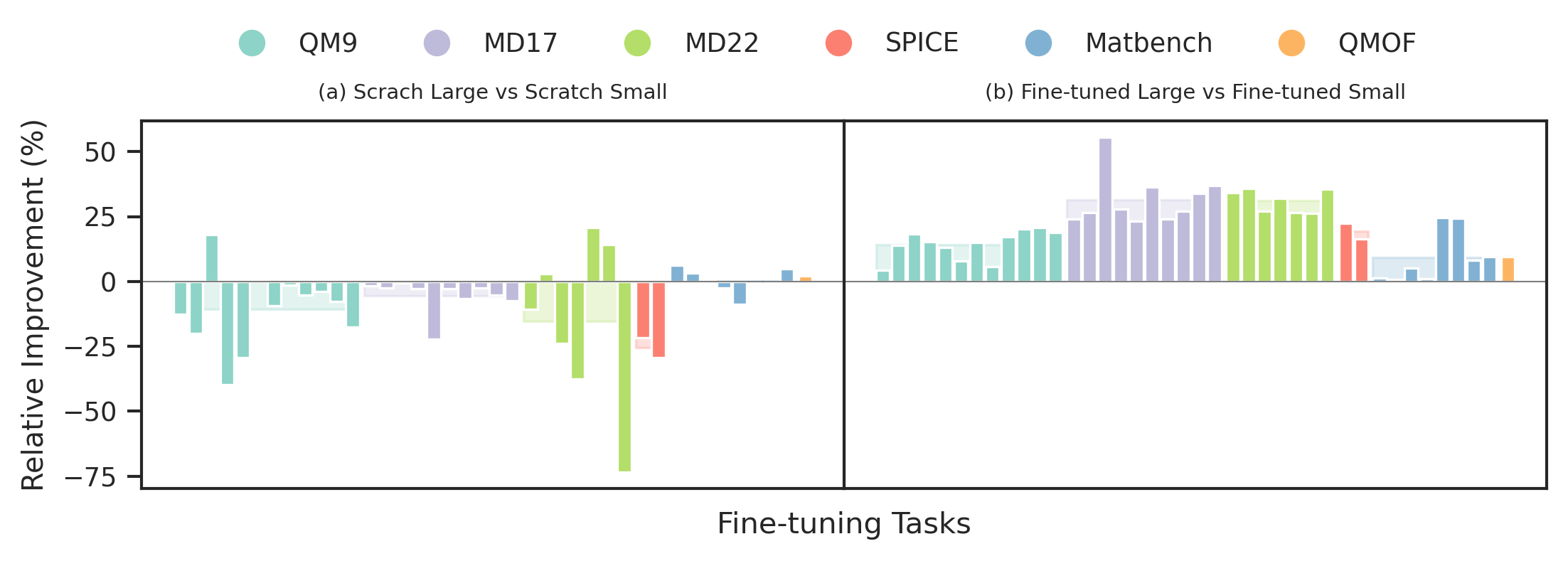
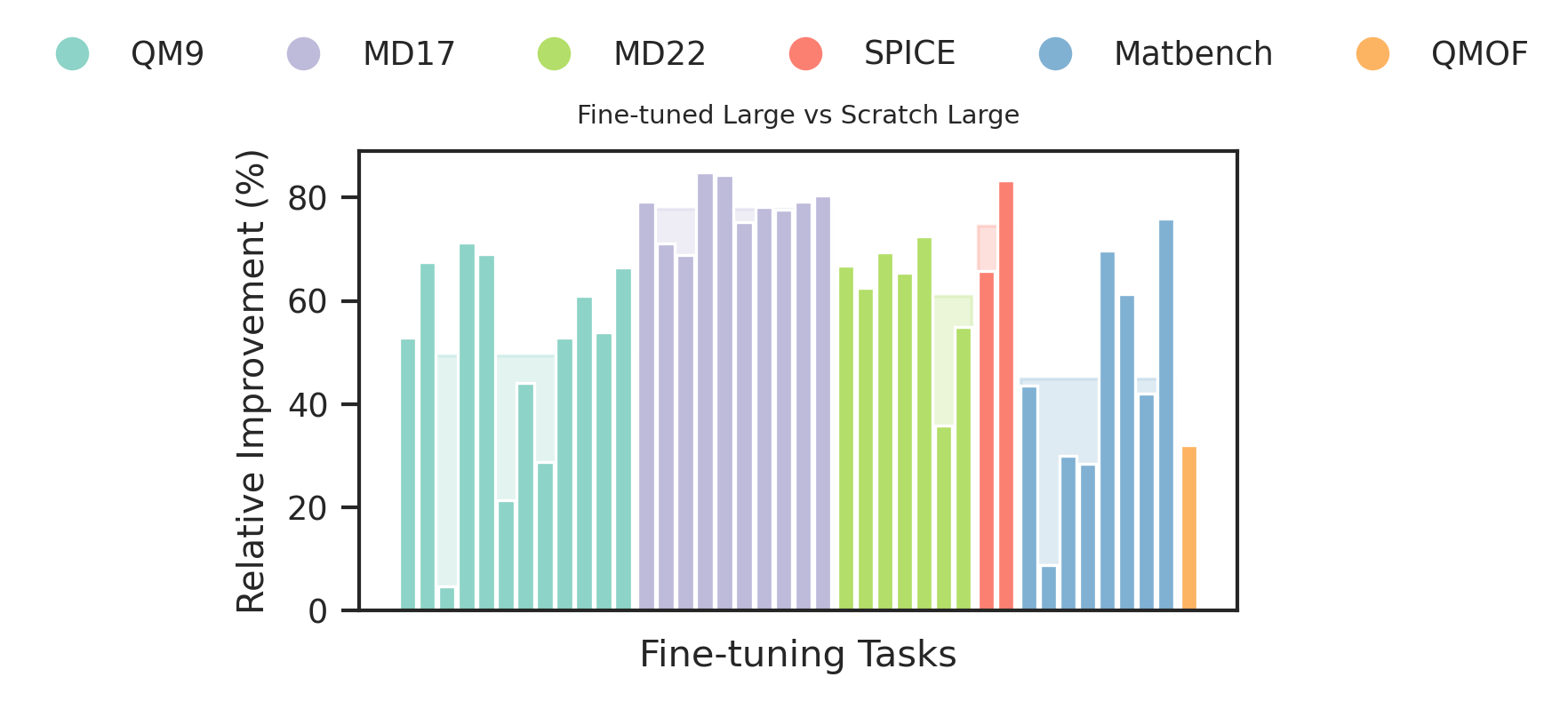
We compare the relative improvement of JMP-L over JMP-S to the relative improvement of GN-OC-L (scratch large model) over GN-OC-S (scratch small model). We observe that the larger models yields 21% average performance improvement in JMP, compared to an 8% reduction in performance in the scratch setting. This demonstrates that JMP enables training larger models without sacrificing performance.
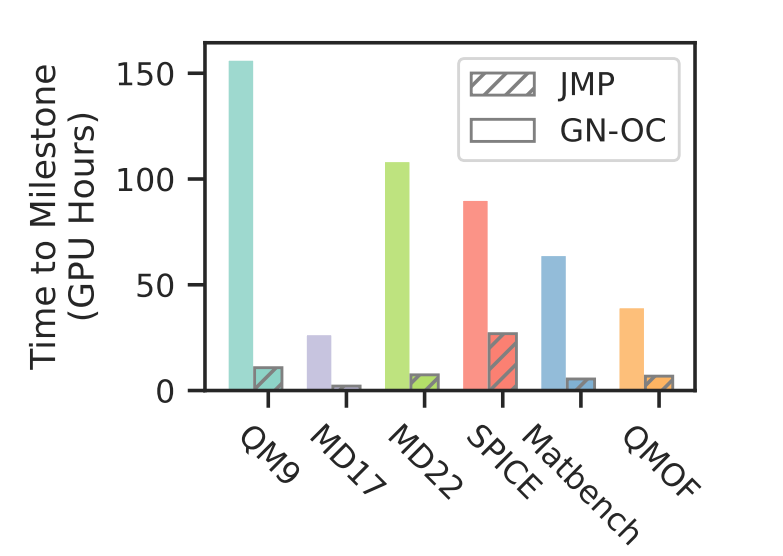
While JMP's pre-training is computationally expensive, taking around 34,400 GPU hours, this upfront cost is recovered by enabling over 12x faster fine-tuning compared to training models from scratch.
We visualize the learned representations in JMP using t-SNE. Each layer of the model is visualized, showing how the representations evolve as the model processes the input data across each GemNet layer.
@article{shoghi2023molecules,
title={From molecules to materials: Pre-training large generalizable models for atomic property prediction},
author={Shoghi, Nima and Kolluru, Adeesh and Kitchin, John R and Ulissi, Zachary W and Zitnick, C Lawrence and Wood, Brandon M},
journal={arXiv preprint arXiv:2310.16802},
year={2023}
}