From Molecules to Materials
Pre-training Large Generalizable Models for Atomic Property Prediction
*Corresponding author
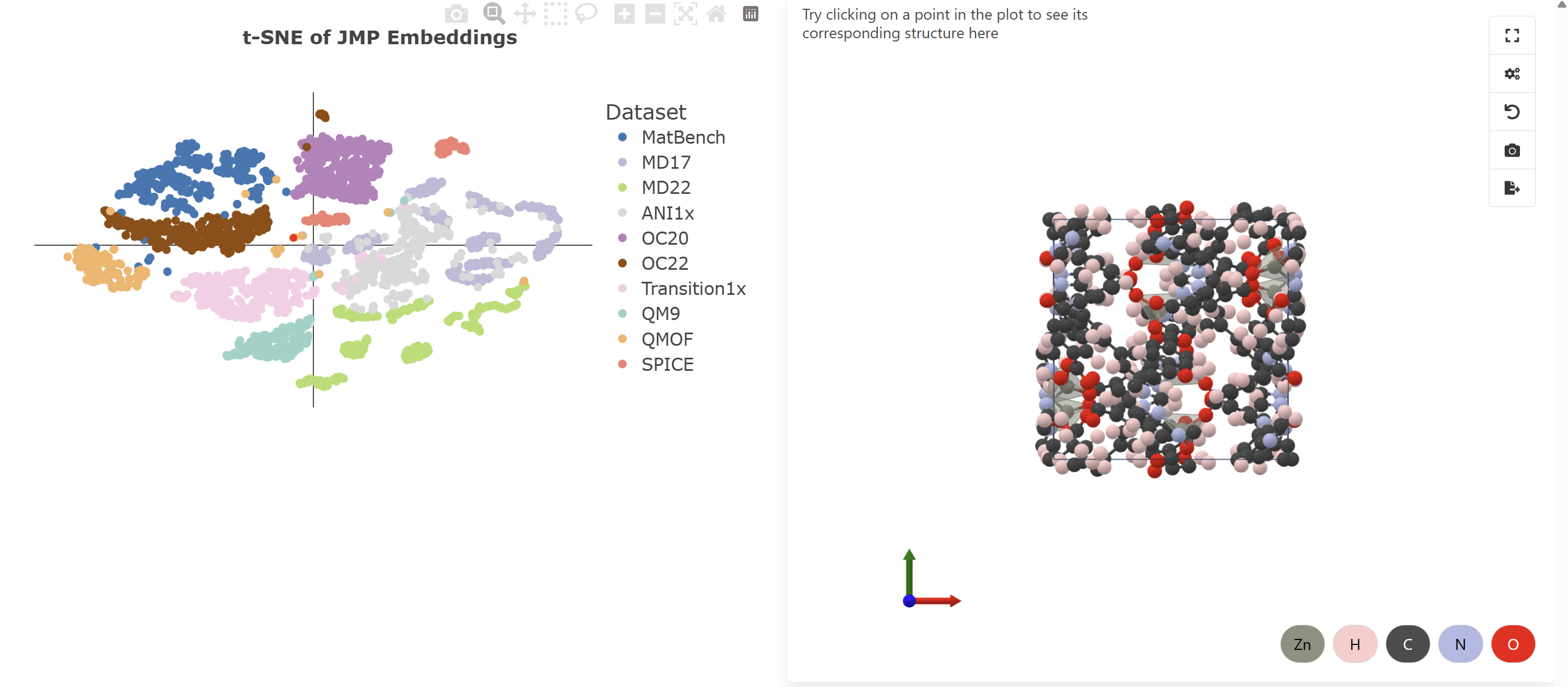
t-SNE Visualization: JMP embeddings across 10 datasets showing unified atomic representations. Explore interactively in Colab →
Abstract
Atomic property prediction plays a central role in computational chemistry and materials science. However, developing accurate and efficient predictive models remains challenging due to the complex interactions between atoms and the large data requirements for training robust models.
We propose Joint Multi-domain Pre-training (JMP), a supervised pre-training strategy that simultaneously trains on multiple datasets from different chemical domains, treating each dataset as a unique pre-training task within a multi-task learning framework.
Our combined pre-training dataset consists of approximately 120M systems from OC20, OC22, ANI-1x, and Transition-1x. We benchmark JMP on a diverse set of downstream tasks across small molecules, large molecules, and materials, evaluating fine-tuned models on QM9, rMD17, MD22, SPICE, MatBench, and QMOF datasets.
JMP achieves state-of-the-art results on 34 out of 40 downstream benchmarks. In addition, our work highlights the value of pre-training strategies that utilize diverse data to achieve generalizable representations in atomic property prediction.
Joint Multi-domain Pre-training
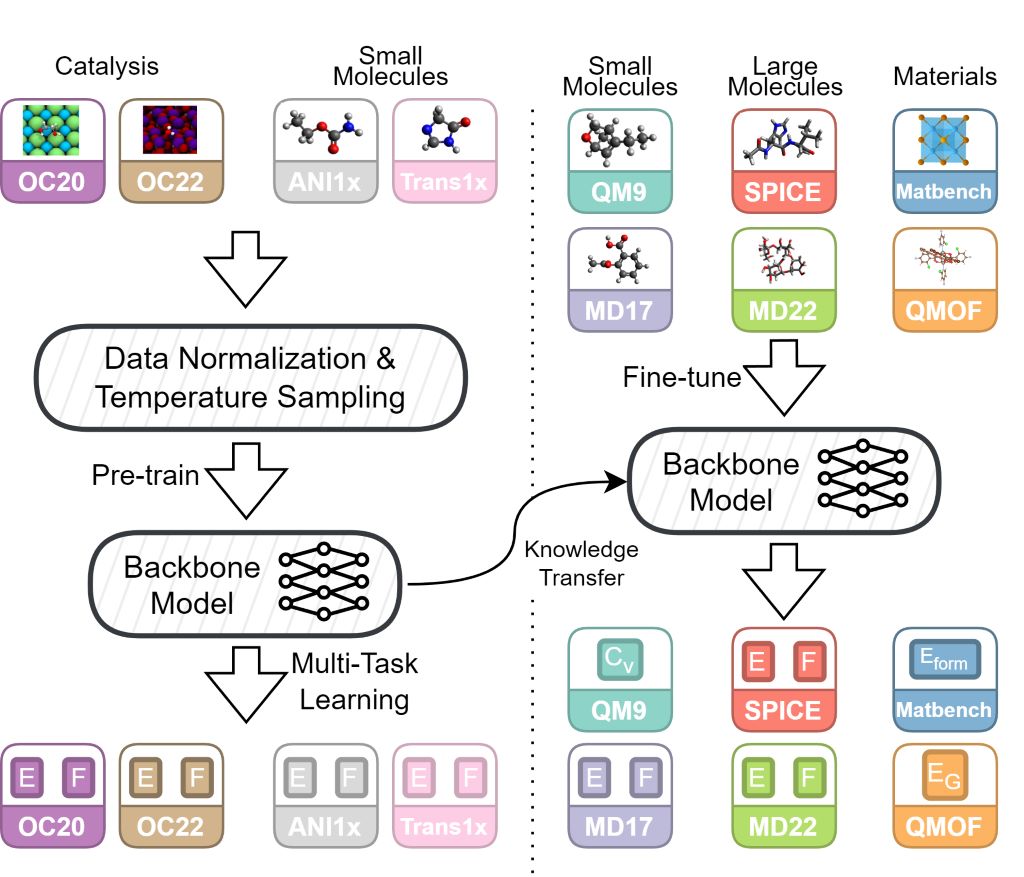
Figure 1: Overview of JMP showing joint pre-training on diverse datasets followed by efficient fine-tuning.
Pre-training
JMP pre-trains on approximately 120 million systems from four diverse datasets, learning unified atomic representations across molecular and materials domains.
Catalyst relaxation trajectories
Oxide catalyst relaxations
Small molecule MD simulations
Reaction transition states
Fine-tuning
Pre-trained JMP models can be efficiently fine-tuned on downstream tasks with minimal data, achieving state-of-the-art on 34/40 benchmarks.
QM9, rMD17
MD22, SPICE
MatBench, QMOF
Benchmark Results
JMP achieves state-of-the-art results across diverse benchmarks spanning small molecules, large molecules, and materials.
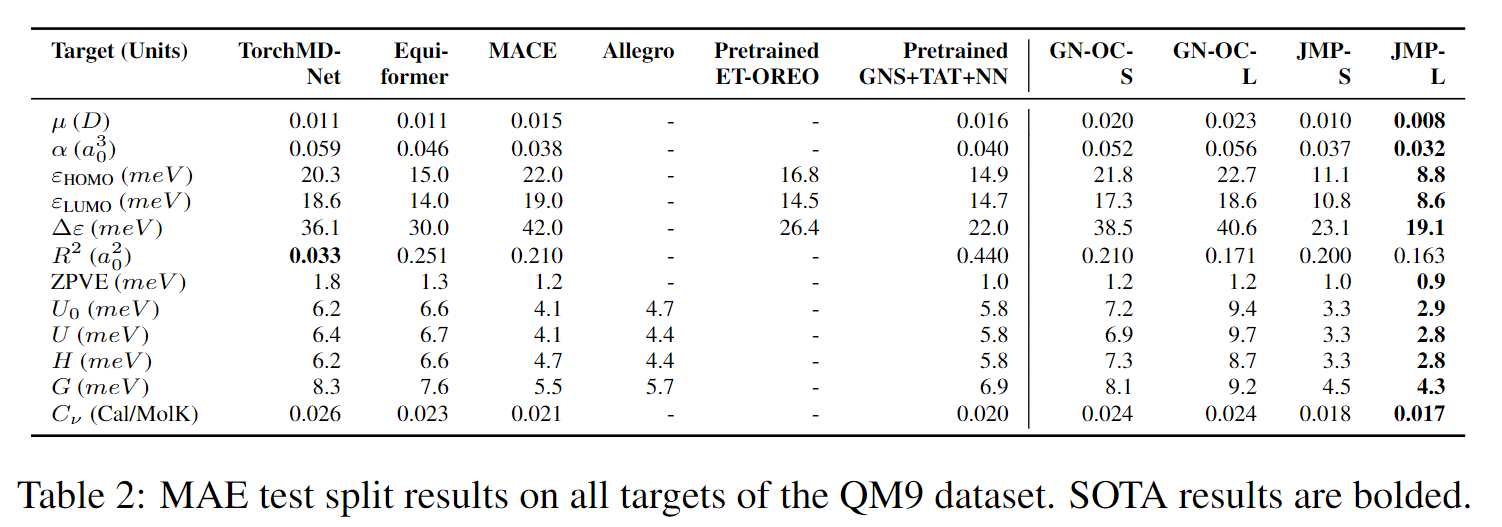
QM9 benchmark results comparing JMP to prior methods.
Key Results
Pre-training Improves Downstream Performance
JMP pre-training yields an average improvement of 59% across downstream tasks compared to training from scratch.

Pre-training Enables Larger Models
Pre-training allows effective scaling to larger models (JMP-L, 235M parameters) that consistently outperform smaller variants.
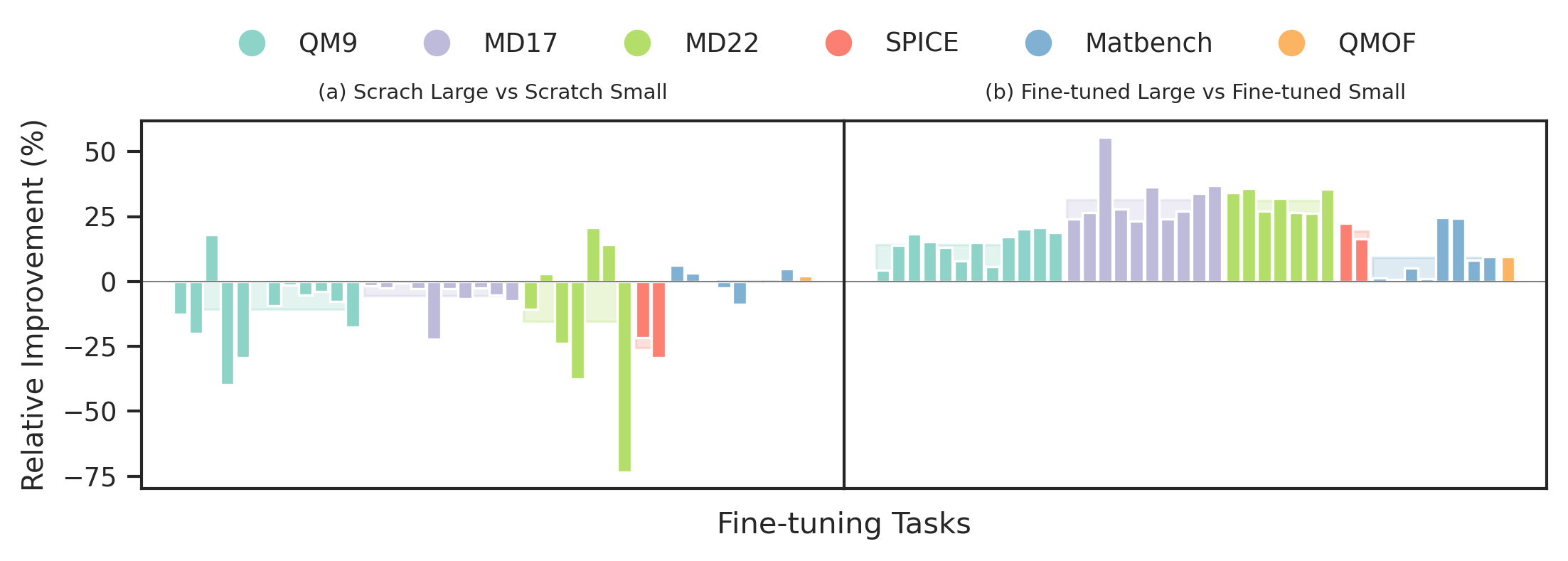
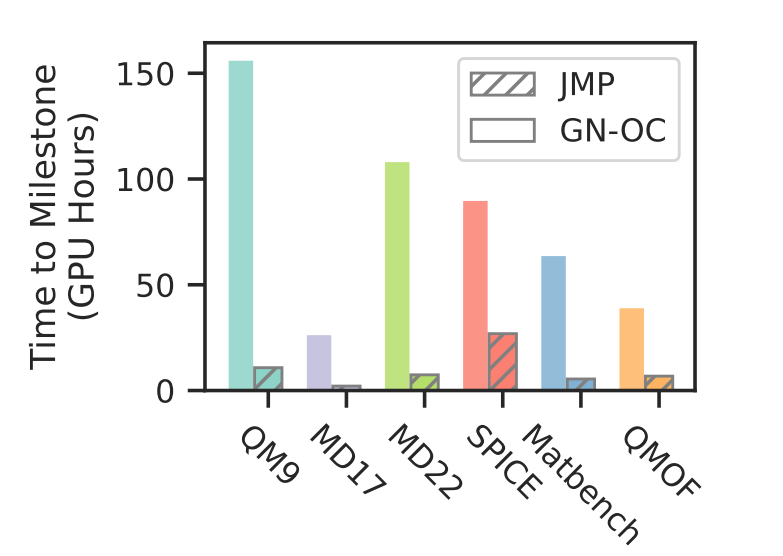
Pre-training Reduces Computational Cost
JMP significantly reduces the GPU hours required for downstream fine-tuning compared to training from scratch.
Per-Layer t-SNE Visualization
Watch how atomic representations evolve through the layers of the pre-trained JMP model, progressively learning more discriminative features.
Layer-by-layer evolution: Atomic embeddings become increasingly domain-specific through deeper layers of the network.
Video Presentation
Watch the ICLR 2024 presentation to learn more about JMP.
Citation
@inproceedings{shoghi2024molecules,
title={From Molecules to Materials: Pre-training Large Generalizable
Models for Atomic Property Prediction},
author={Shoghi, Nima and Kolluru, Adeesh and Kitchin, John R and
Ulissi, Zachary W and Zitnick, C Lawrence and Wood, Brandon M},
booktitle={The Twelfth International Conference on Learning
Representations},
year={2024}
}